Clustered Heatmap
Seaborn basics
2 min read
Published Oct 7 2025
Guide Sections
Guide Comments
seaborn.clustermap() creates a clustered heatmap — a combination of:
- A heatmap (coloured grid of values), and
- Hierarchical clustering on both rows and columns.
It automatically reorders rows and columns to group similar patterns together, and draws dendrograms (tree diagrams) showing the clustering structure.
It’s commonly used in:
- Gene expression analysis
- Feature correlation exploration
- Pattern detection in any tabular numeric data
Syntax:
Parameters:
data= 2D data (DataFrame or matrix)method= Clustering algorithm: "single", "complete", "average", "ward", etc.metric= Distance measure: "euclidean", "correlation", "cityblock", etc.z_score= Normalise rows (0) or columns (1) to z-scoresstandard_scale= Scale rows (0) or columns (1) between 0 and 1cmap= Colour map for the heatmapcenter= Value to centre the colourmapannot= Display numerical values in cellsrow_cluster/col_cluster= Enable/disable clustering on rows or columnsrow_linkage,col_linkage= Precomputed linkage matrices for custom clusteringxticklabels,yticklabels= Show/hide or customise labelsfigsize= Size of the entire clustered plottree_kws= Arguments for dendrogram styling
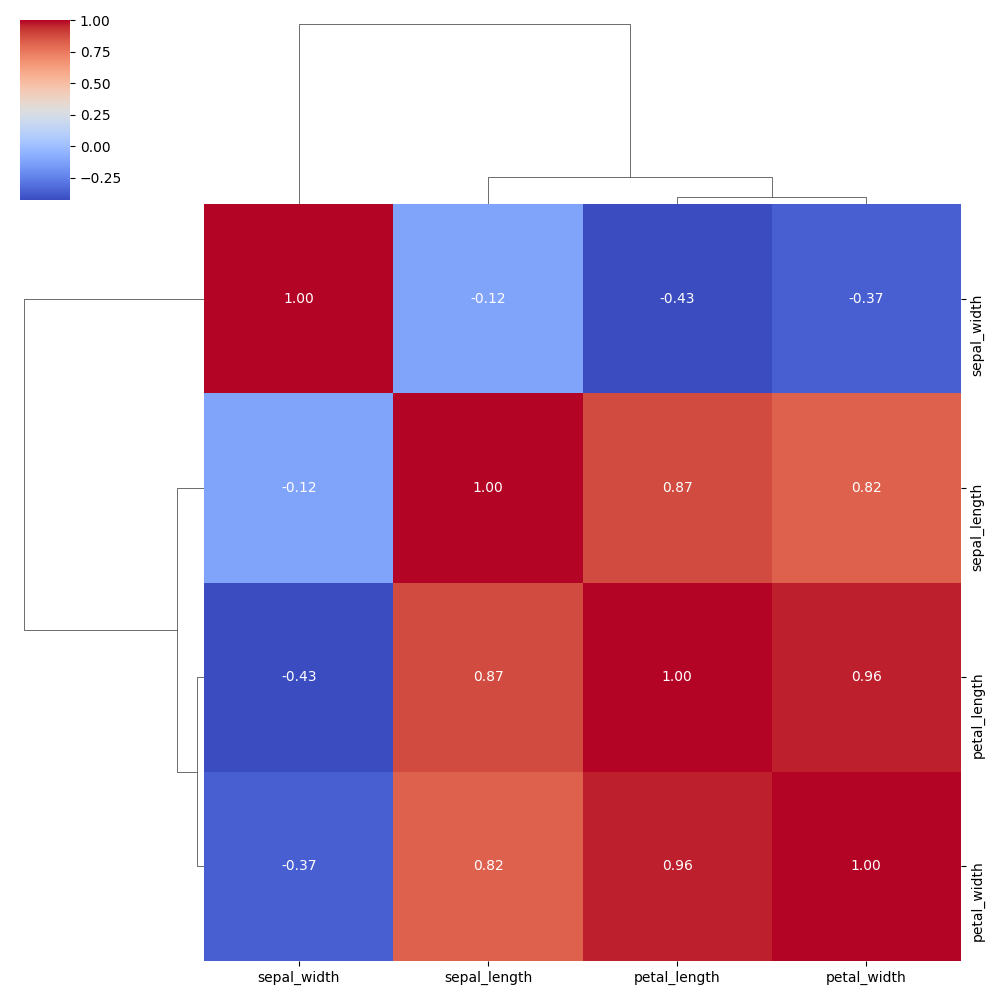
Basic example
Displays a correlation matrix clustered by similarity. Similar variables are grouped together with dendrograms on top and left.
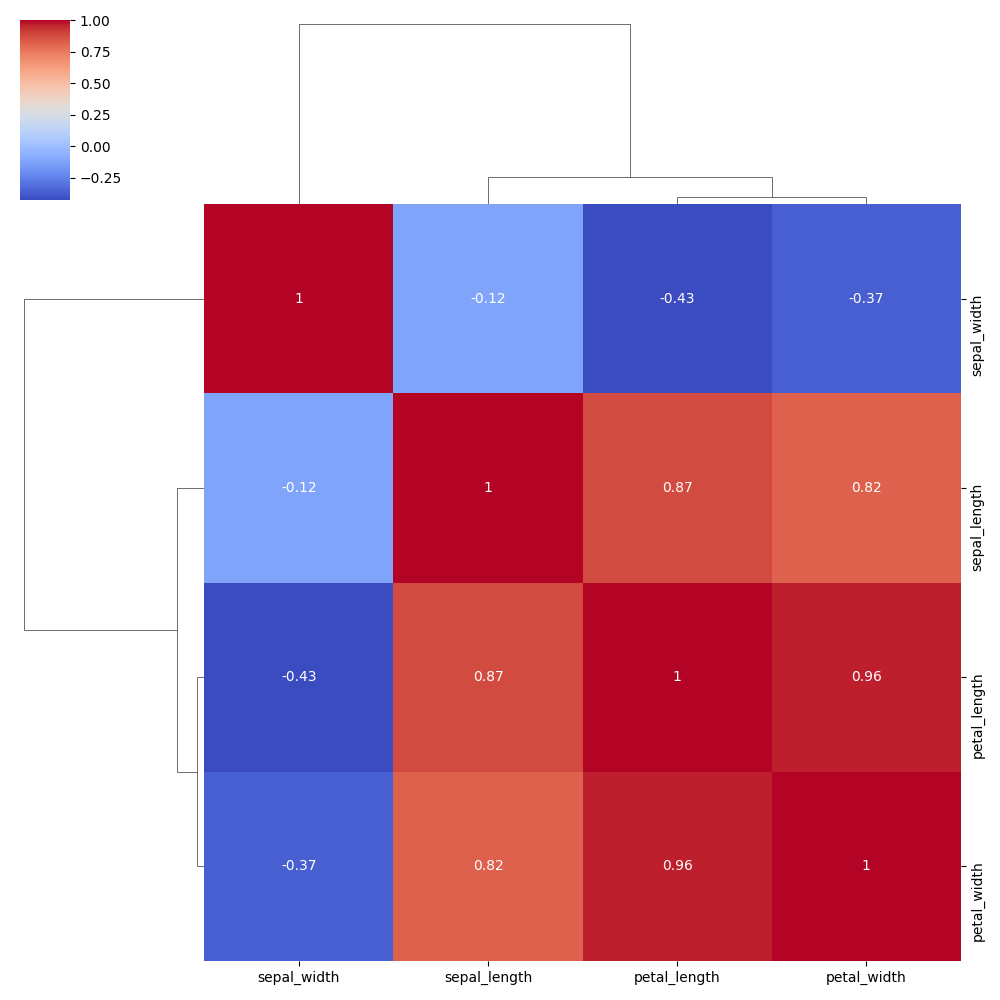
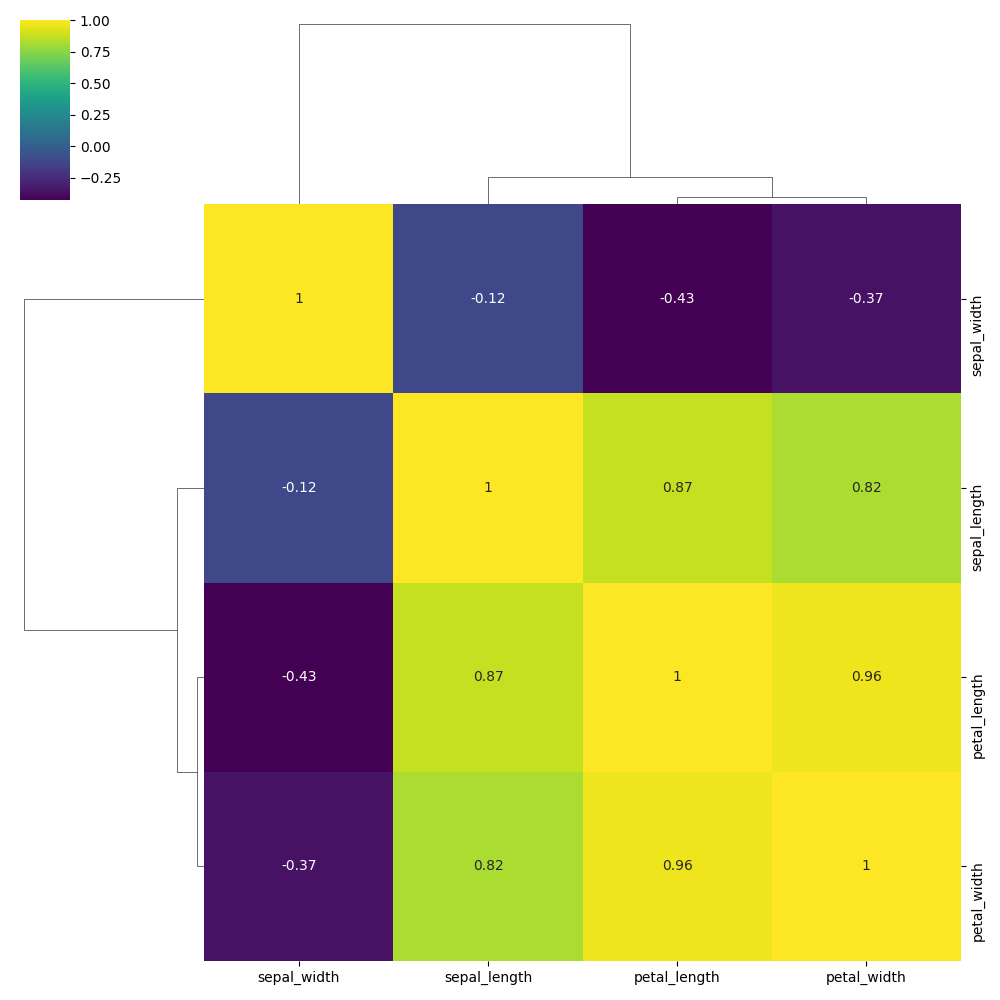
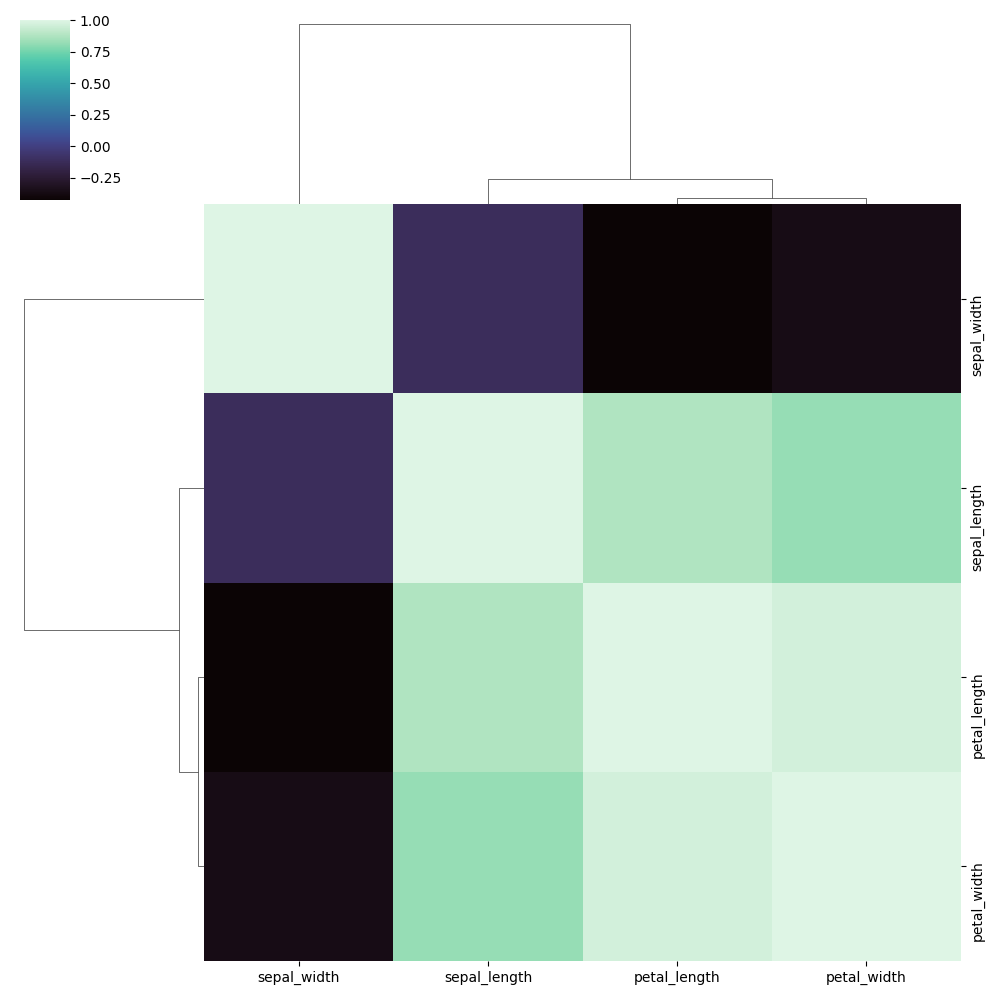
Customise color palette
You can use any Matplotlib or Seaborn colourmap ("mako", "crest", "rocket", etc.).
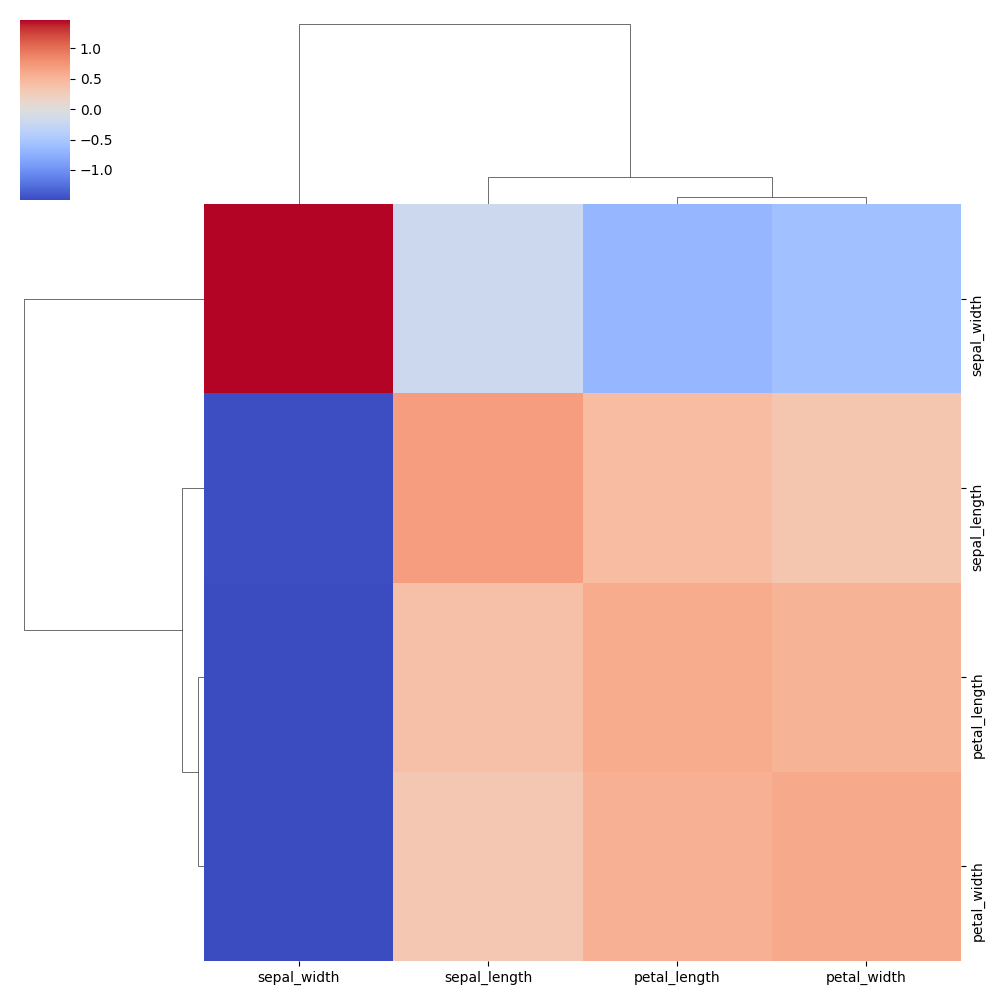
Normalise data (z-score)
Normalises data by rows to z-scores (mean = 0, std = 1). Use z_score=1 to normalise by columns.
Disable clustering
Keeps the data order fixed along the chosen axis.
No Row Clustering:
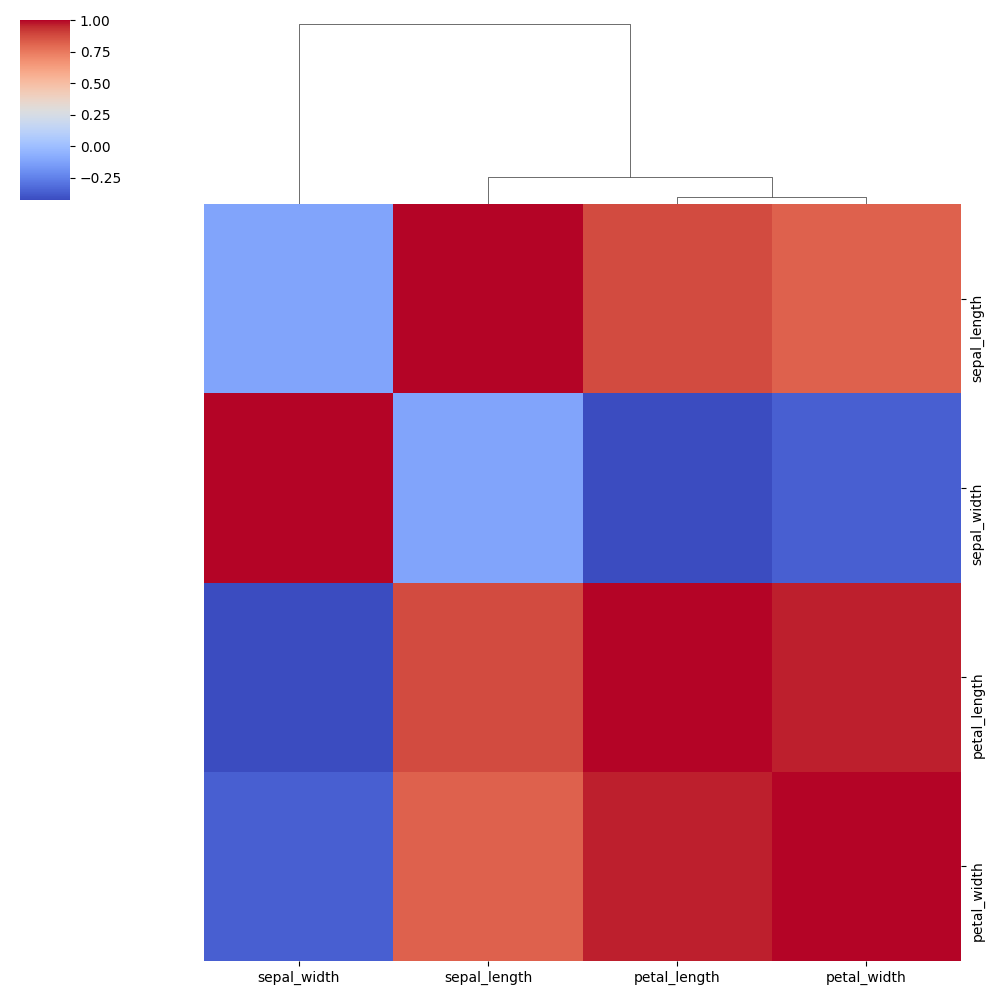
No Column Clustering:
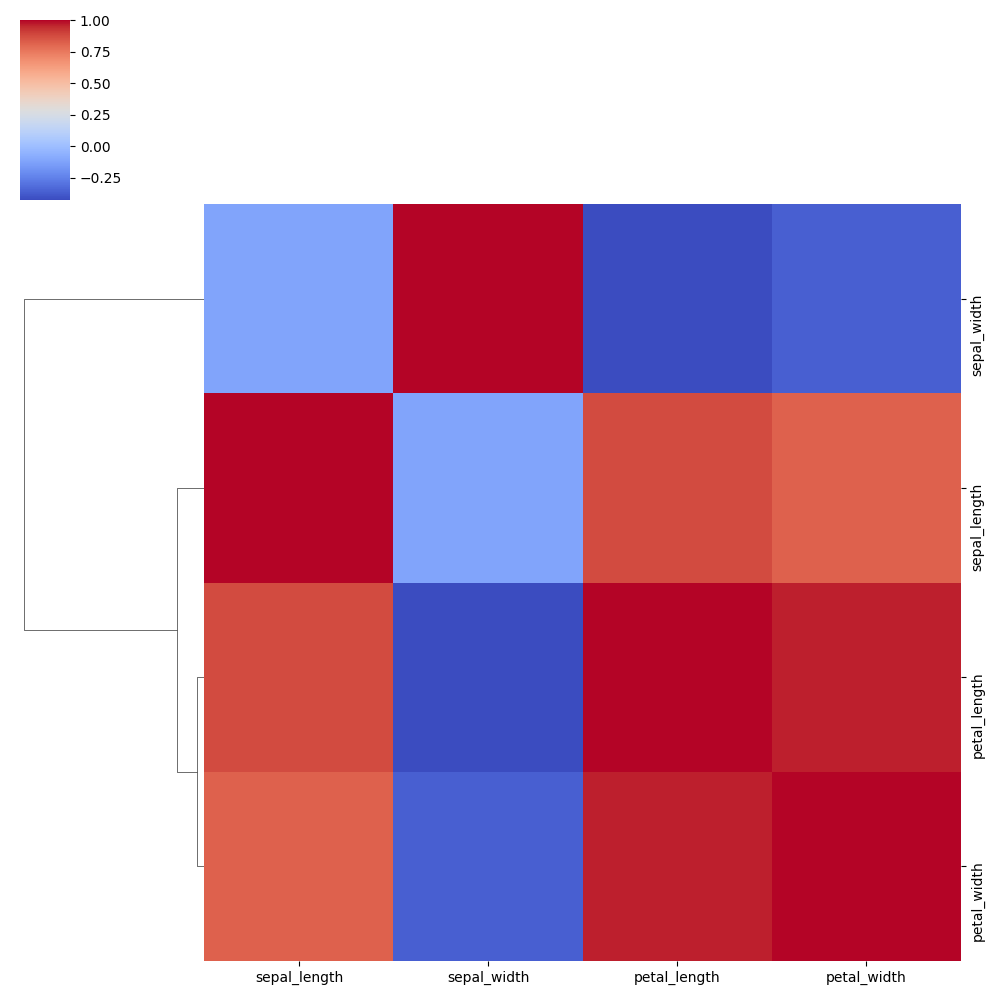
Change clustering method and distance
Controls how clusters are formed:
method: linkage type ("ward","average","complete", etc.)metric: distance measure ("euclidean","cosine","correlation", etc.)
Add annotations
Displays the numeric values inside each cell. Great for small matrices (e.g., correlation heatmaps).